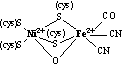Bioinorganic Chemistry of Nickel
Researchers have become increasing interested in the
coordination chemistry of nickel complexes as models for the active sites
in nickel containing enzymes1-3.
This is largely due to the discovery of nickel at the centers of many
important enzymes4-7. There
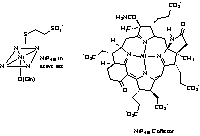
are six nickel enzymes discovered so far, they are: urease8, NiFe
hydrogenases, methyl coenzyme M reductase9, carbon monoxide dehydrogenase10,
acetyl coenzyme A synthase11 and more recently nickel superoxide
dismutase (NiSOD)12, 13. Additionally, there are several proteins
required for the delivery and assembly of the nickel ions into the active
sites of some of these proteins. For example, the nickel-uptake system in Escherichia coli
consists of five proteins, NikABCDE.
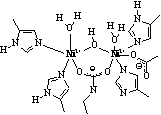
Recent spectroscopic studies of NikA suggest that the nickel
site is six coordinate and comprised of five oxygen and/or nitrogen donors,
and a single sulfur donor14-16.
There are additional E. coli enzymes in which nickel is important
such as peptide deformylase and glyoxalase I which catalyze the deformylation
of nascent polypeptides, and the isomerization reaction of the hemiacetal
to the thioester of d-lactate formed from the reaction of pyruvaldehyde and
glutathione respectively17. Since
1995, the structural biology of nickel proteins has greatly expanded largely
due the X-ray structures of urease18, NiFe hydrogenase19,
and methyl coenzyme M reductase20. Many of the enzymatic reactions which occur
in these centers are primarily due to the redox activity of the nickel atoms
in the enzyme’s active site (Table
(in pdf format)). In fact the proposed mechanisms of NiFe hydrogenase21,
methyl-CoM reductase22,23 and NiSOD12, 13 involves reduced
(Nio,Ni+) and or oxidized (Ni3+) forms of
nickel. In the case of Methyl-CoM
reductase, and NiFe Hydrogenase a Ni(III)-Ni(II)-Ni(I) cycle has
been proposed as part of the enzymatic cycle24,25. It is important to note that in general, the
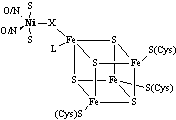
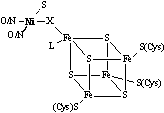
nickel sites in the redox enzymes are dominated by cysteinate ligation whereas
the nickel sites in nonredox, hydrolytic enzymes and transport proteins are
dominated by O(N)-donor ligands. Therefore,
the trend is for biological redox active nickel centers to be closely associated
with sulfur-ligation whereas those with nonredox roles lack Ni-S bonds.
It
is difficult to achieve this Ni(III)-Ni(II)-Ni(I)
redox cycle in small molecule chemistry mainly due to the fact that the
coordination environments which tend to stabilize Ni+ are much
different from those that stabilize Ni3+ 26-28. Nickel centers with reversible Ni(II)/Ni(I) and Ni(III)/Ni(II) couples
and low Ni(III)/Ni(II) potentials are crucial to the activity of these nickel
redox enzymes. The importance of these
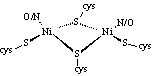
characteristics has lead to an increased interest in the synthesis of Ni(II)
complexes with mixed N/S donors as structural, spectroscopic and redox models
of the active sites of these redox enzymes29, 30.
(1) Halcrow, M. A.; Christou, G. Chem. Rev. 1994 94, 2421.
(2) Stavropoulos, P.; Muetterties, M. C; Carrié, M.; Holm, R. H. J. Am. Chem. Soc. 1991, 113, 8485.
(3) Tucci, G. C.; Holm, R. H. J. Am. Chem. Soc. 1995, 117, 6489.
(4) Tommasi, I.; Aresta, M.; Giannoccaro, P.; Quaranta, E.; Fragale, C. Inorg. Chim. Acta. 1998, 272, 38.
(5) Cammack, R.; vanVlient, P. V., In Bioinorganic Catalysis, Reedijk,
J.; Bouwman, E., Eds., Marcel Dekker, New York, 1999, Chapter 9,
pgs. 231-233.
(6) Ermler, U.; Grabarse, W.; Shima, S.; Goubeaud, M, Thauer, R. K.
Curr. Op. Struct. Biol. 1998, 8, 749.
(7) Dole, F.; Medina, M.; More, C.; Cammack, R.; Bertrand, P.;
Guigliarelli, B. Biochem. 1996, 35, 16399.
(8) Andrews, R. K.; Blakeley, R. L.; Zerner, B. In The Bioinorganic
Chemistry of Nickel, Lancaster, J. R., Ed., VCH Publishers, New
York, 1988, Chapter 7 pgs. 141-165.
(9) Telser, J. Struct. Bonding 1998, 91, 31.
(10) Ralston, C. Y.; Wang, H.; Ragsdale, S. W.; Kumar, M.; Spangler, N
.J.; Ludden, P. W.; Gu, W.; Jones, R. M.; Patil, D. S.; Cramer, S. P.
J. Am. Chem. Soc. 2000, 122, 10553.
(11) Ragsdale, S. W.; Kumar, M. Chem. Rev. 1998, 96, 2515.
(12) Youn, H. -D.; Kim, E. -H.; Roe, J. -H.; Kang, S. -O.; Biochem. J.
1996, 318, 889.
(13) Choudhury, S. B.; Lee, J. –W.; Davidson, G.; Yim, Y. -I.; Bose, K.;
Sharma, M. L.; Kang, S. -O.; Cabelli, D. E.; Maroney, M. J.
Biochemistry 1999, 38, 3744.
(14) (a) Eitinger, T. Arch. Microbiol. 2000, 173, 1. (b)Allan, C. B.; Wu,
L.-F.; Gu, Z.; Choudhury, S. B.; Al-Mjeni, F.; Sharma, M. L.; Mandrand-Berthelot, M-A.; Moroney, M. J. Inorg. Chem. 1998, 37, 5952.
(15) de Pina, K.; Navarro, C.; McWalter, L.; Boxer, D. H.; Price, N. C.;
Kelly, S. M.; Mandrand-Berthelot, M.-A.; Wu, L. –F. Eur. J. Biochem. 1995, 227, 857.
(16) Charon, M. –H; Wu, L. –F.; Piras, C.; de Pina, K.; Mandrand-
Berthelot, M.-A.; Fontecilla-Camps, J. C. J. Mol. Biol. 1994, 243, 353.
(17) Davidson, G.; Clugston, S. L.; Honek, J. F.; Maroney, M. J. Inorg.
Chem. 2000, 39, 2962.
(18) Jabri, E.; Carr, M. B.; Hausinger, R. P.; Karplus, P. A. Science 1995,
268, 998.
(19) Volbeda, A.; Charon, M. H.; Piras, C.; Hatchikian, E. C.; Frey, M.;
Fontecilla-Camps, J. C. Nature 1995, 373, 580.
(20) Ermler, U.; Grabarse, W.; Shima, S.; Goubeaud, M.; Thauer, R. K.
Science 1997, 278, 1457.
(21) Kaim, W.; Schwederski, B.; Bioinorganic Chemistry: Inorganic
Elements in the Chemistry of Life: An Introduction and Guide, 1991, Wiley, Sussex, Chapter 9, pgs 175-178.
(22) Jaun, B. Helv. Chim. Acta 1990, 73, 2209.
(23) Berkessel, A. Bioorg. Chem. 1991, 19, 101.
(24) Cammack, R.; Rao, K. K.; Serra, J.; Llama, M. J. Biochemie 1986,
68, 93.
(25) Cammack, R. Adv. Inorg. Chem. 1988, 32, 297.
(26) Sabatini, S.; Fabbrizzi, L. Inorg. Chem. 1979, 18, 438.
(27) Barefield, E. K.; Freeman, G. M.; Van Derveer, D. G. Inorg. Chem.
1986, 25, 552.
(28) deVries, N.; Reedijk, J. Inorg. Chem. 1991, 30, 3700.
(29) Musie, G.; Farmer, P. J.; Tuntulani, T.; Reibenspies, J. H.;
Darensbourg, M. Y. Inorg. Chem. 1996, 35, 2176.
(30) Goodman, D. C.; Buonomo, R. M.; Farmer, P. J.; Reibenspies, J. H.;
Darensbourg, M. Y. Inorg. Chem. 1996, 35, 4029.
|
Nickel Enzymes (active sites,
reactions and biological distribution) |
||
Enzyme
|
Ni-center |
Reaction and Distribution |
Urease
|
|
CO(NH2)2 à NH3 + CO2 + HNCO Bacteria and Plants
|
|
NiFe Hydrogenase* |
|
2H+(aq)
+ 2e- ¾ H2 Bacteria |
|
Methyl-CoM reductase* |
|
MethanogenicBacteria |
|
CO dehydrogenase |
|
CO + H2O à CO2 + 2H+ + 2e- Photosynthetic Bacteria |
|
Acetyl CoA synthase |
|
CO2 + [CH3]-Co(III)balamin +SCoA +2e- à CH3C(O)ScoA+ Co(I)balamin Methanogenic and
Acetogenic Bacteria |
|
Nickel Superoxide Dimutase* |
|
2 O2·- + 2H+ à O2 + H2O2 Streptomyces |